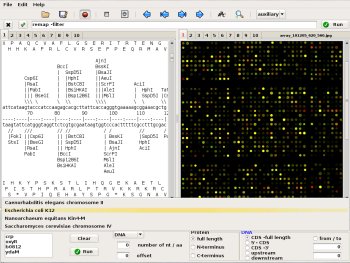
SeqFreed is a bioinformatics desktop. It provides efficient and user-friendly access to sequence data and gene features. Its customizable command line interface runs locally installed bioinformatic tools in a convenient way. Seqfreeds flexible display of either text or images in different page modes enhances ease and productivity of routine molecular biology analysis. SeqFreed is written in Python using GTK+ and runs on Linux.
SeqFreed 0.01 released - 02 Apr 2006
Welcome.This is the first release of SeqFreed, a bioinformatics desktop.
The basic design of SeqFreeds webpage was copied from the Comix site (comix.sourceforge.net) with the permission of Pontus Ekberg. Thank you, Pontus!
Features
- -- Sequence and feature extraction from multiple GenBank files.
- -- Database like access to sequence data:
- Protein or DNA.
- Full length sequences or subsequences.
- Coding or noncoding regions.
- Sequence access by gene name, locus tag or absolute coordinates.
- -- Fast, index-driven lookup.
- -- Batch lookup: arbitray number of gene queries processed at once.
- -- Customizable command line (history) with entry completion.
- -- Page content is piped as input to the invoked application.
- -- Results are directly read from stdin and displayed as either text or image.
- -- Single page, double page and "full screen" mode.
- -- 20 notebook pages for text or image display.
- -- Text font size scalable.
- -- Image size scalable.
- -- Convenient "single-click-copy" and "single-click-append".
- -- Page lock prevents accidental overwrite.
- -- Images saved as PNG.
- -- User definable auxiliary pages.